
Low-coverage sequencing in a deep intercross of the Virginia body weight lines provides insight to the polygenic genetic architecture of growth: novel loci revealed by increased power and improved genome-coverage - ScienceDirect

The minor allele frequency (x-axis distribution of collapsed SNPs using

The construction of a haplotype reference panel using extremely low coverage whole genome sequences and its application in genome-wide association studies and genomic prediction in Duroc pigs - ScienceDirect
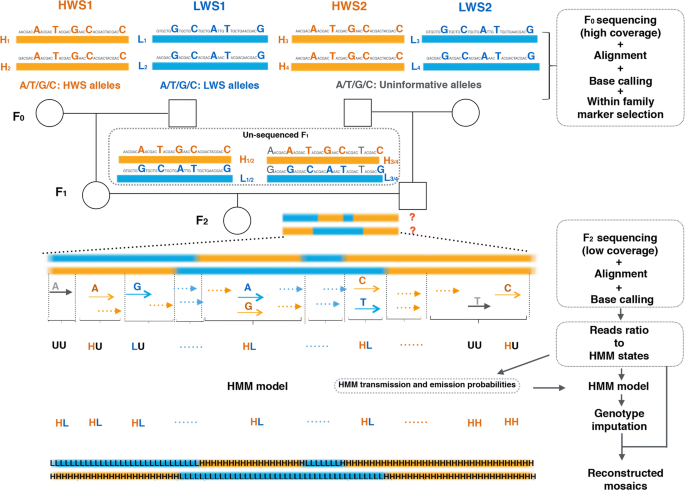
PDF) Genotyping by low-coverage whole-genome sequencing in intercross pedigrees from outbred founders: a cost-efficient approach
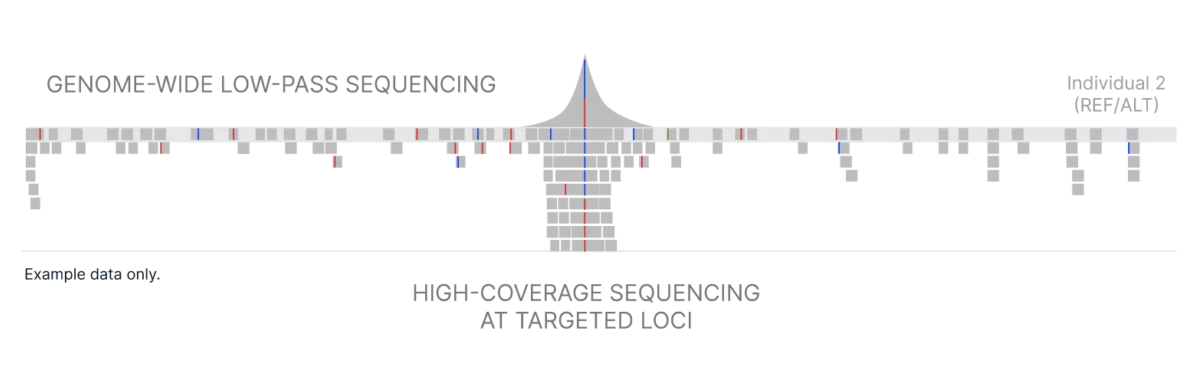
Introducing InfiniSEEK - Low-pass Genotyping Plus Target Capture in One Assay - Gencove
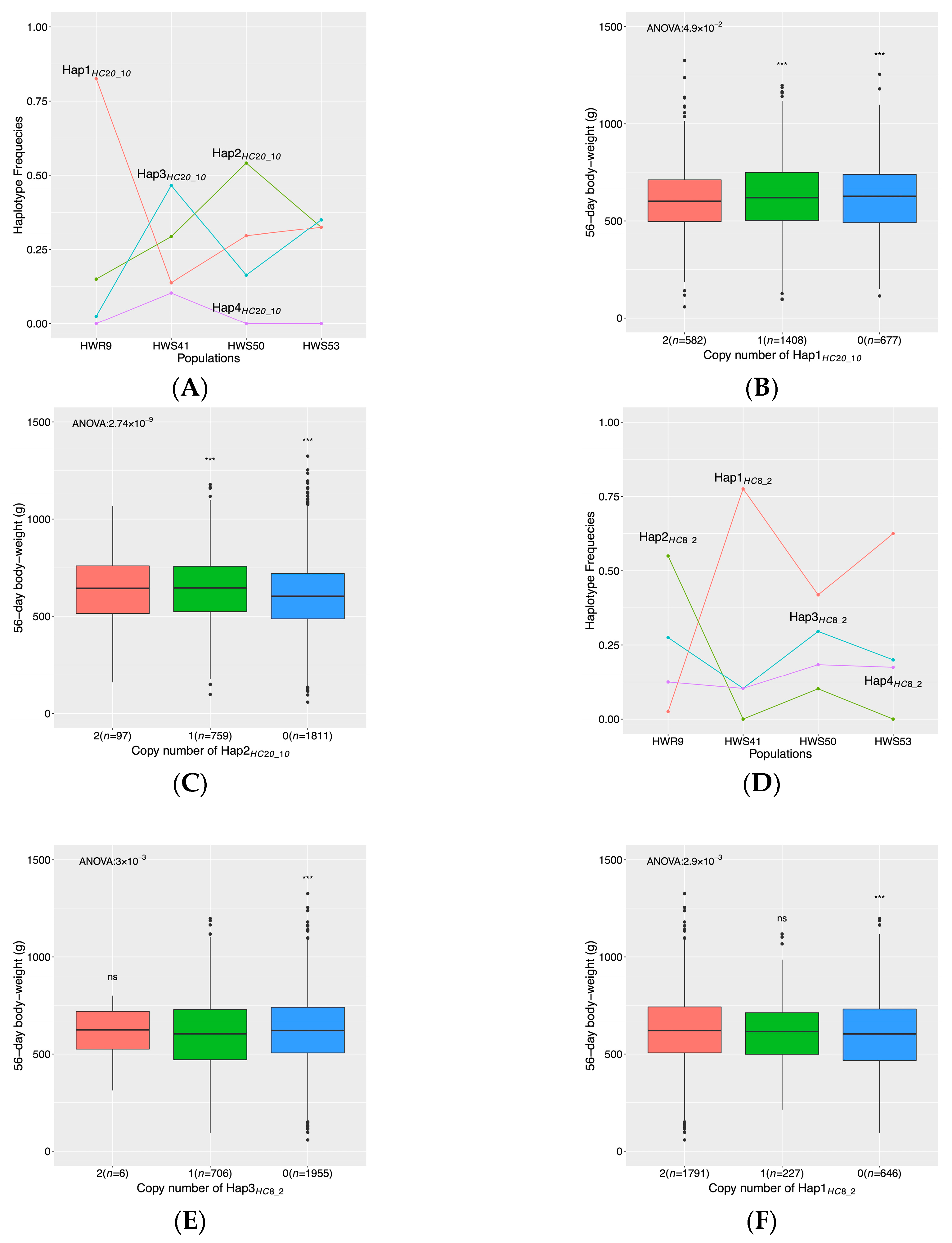
Genes, Free Full-Text

Insects, Free Full-Text

Figure S3. Ilustration of the relationship between sequencing coverage

Assessment of the performance of different imputation methods for low-coverage sequencing in Holstein cattle - ScienceDirect

Identification of RP1 as the genetic cause of retinitis pigmentosa in a multi-generational pedigree using Extremely Low-Coverage Whole Genome Sequencing (XLC-WGS) - ScienceDirect

PDF] Accurate Genotype Imputation in Multiparental Populations from Low-Coverage Sequence

Accelerated deciphering of the genetic architecture of agricultural economic traits in pigs using a low-coverage whole-genome sequencing strategy. - Abstract - Europe PMC

Characterization of a haplotype-reference panel for genotyping by low-pass sequencing in Swiss Large White pigs, BMC Genomics

Diagram of process for imputing missing genotypes. We first construct a

CRISPR/Cas9-based repeat depletion for the high-throughput genotyping of complex plant genomes







